Utilizing Next-Generation Sequencing and Bioinformatics to Enhance Genomic Diagnostics.
My expertise in Next Generation Sequencing (NGS) developed during my postdoctoral training at the University of Wisconsin, where I designed sequencing strategies and executed advanced sequencing technologies including Illumina, Nanopore, and PacBio. This experience was enriched by my role at ATCC, where I led the development of NGS standards and bioinformatics tools for microbiome research, producing over 20 microbiome standards and characterizing more than 1000 microbial genomes. Additionally, I received exceptional training in cloud and high-performance computing as well as programming in Linux Bash, R, and Python.
Over the past decade, I've managed the full NGS workflow from sample preparation to data analysis, contributing to drug discovery and the development of NGS products for microbiome and microbial genome analysis. Today, I apply this extensive NGS expertise at EDAN Diagnostics and GenMol X to enhance diagnostic precision and offer groundbreaking genomic insights.


Next Generation Sequencing (NGS)
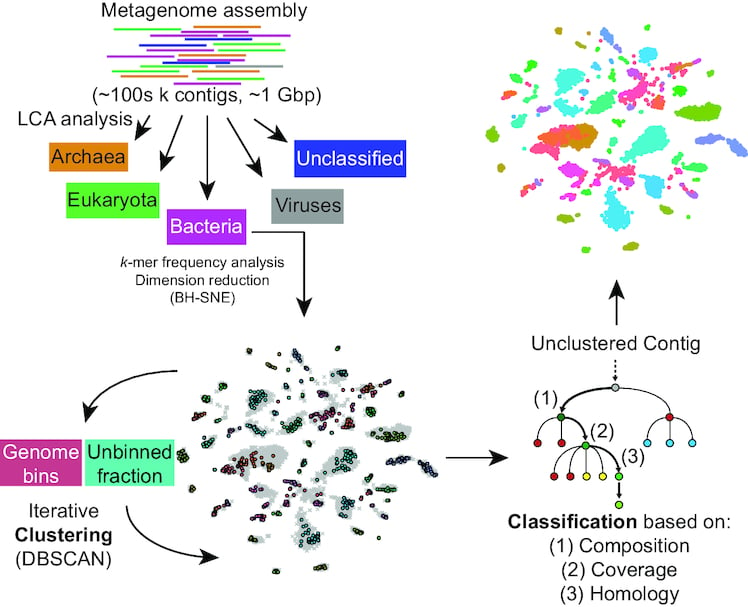
SOME EXAMPLES OF MY NGS WORK